Difference between revisions of "REProcess Export To XGMML File"
(→Result) |
Anote2Wiki (talk | contribs) |
||
| Line 2: | Line 2: | ||
== Operation == | == Operation == | ||
| − | This operation allows | + | This operation allows you to export the entities/relations contained in an REProcess to a network/ graph file in the '''XGMML''' format. |
| + | This file can be afterwards be opened in network visualization and topology analysis such as [http://www.cytoscape.org/ Cytoscape] | ||
| + | |||
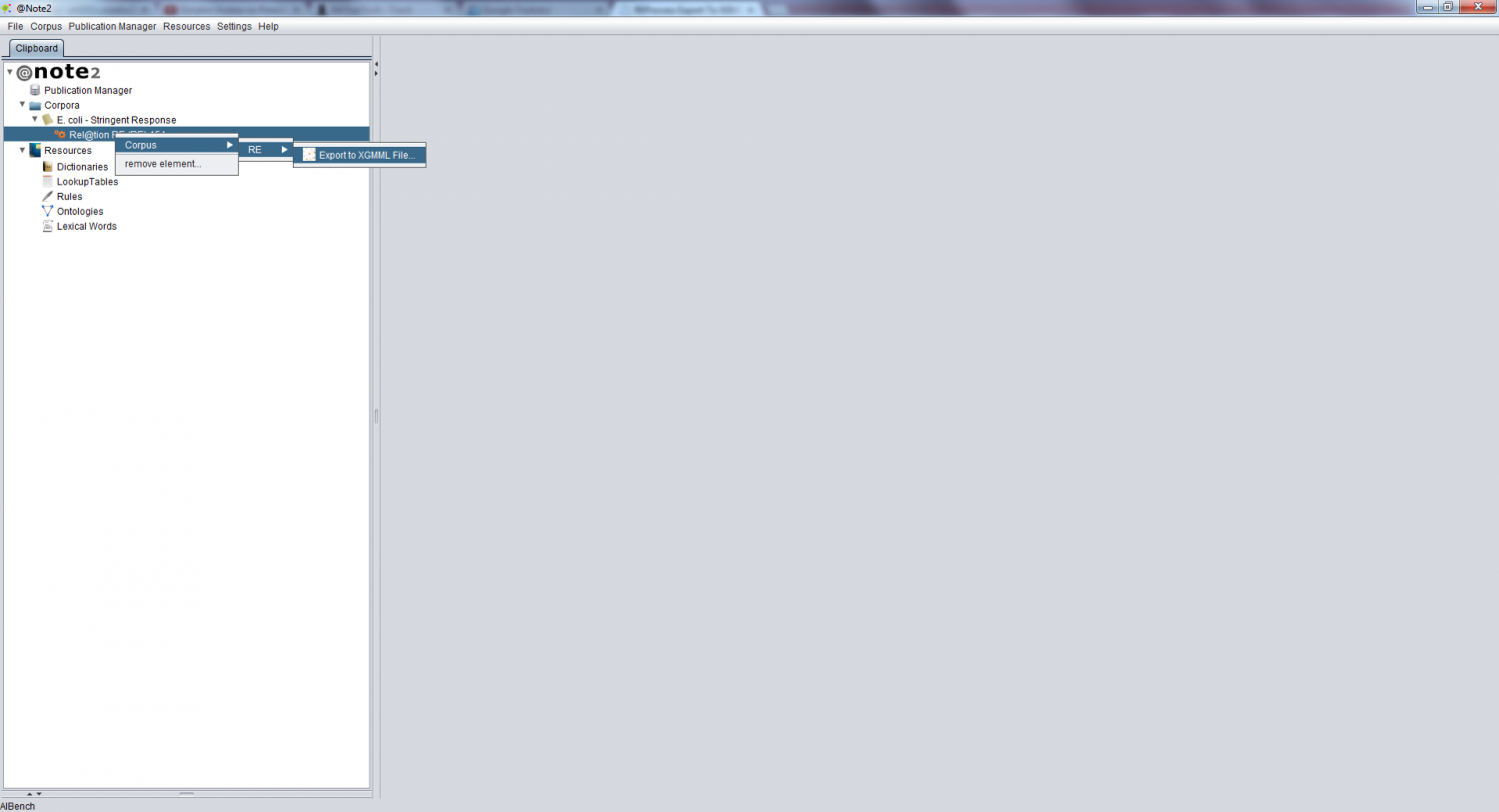
| + | To perform this operation, you need to first select the desired RE Process and right click to select '''''Corpus -> RE -> Export To File XGMML''''' | ||
| − | |||
[[File:RE_To_XGMML_1.png|center|1500px]] | [[File:RE_To_XGMML_1.png|center|1500px]] | ||
| − | + | ||
| + | A wizard will be launched for the configuration of the export process. | ||
== Entity Selection - Classes (Nodes) == | == Entity Selection - Classes (Nodes) == | ||
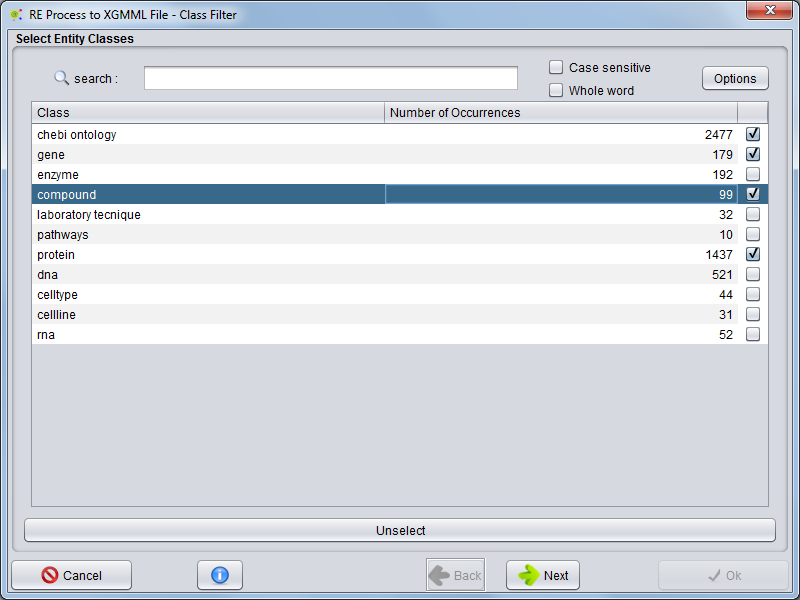
| − | In the first panel, | + | In the first panel, you can choose which entities groups (classes) will be exported to the network file. |
| + | |||
[[File:RE_To_XGMML_2.png|center|800px]] | [[File:RE_To_XGMML_2.png|center|800px]] | ||
| + | |||
== Relation Selection == | == Relation Selection == | ||
| − | + | In the next panel, you can filter the relationships to be included in the network to export. Here, you can selecting which features the relationships need to have (e.g. in terms of directionality, cardinality and polarity). | |
=== Cardinality === | === Cardinality === | ||
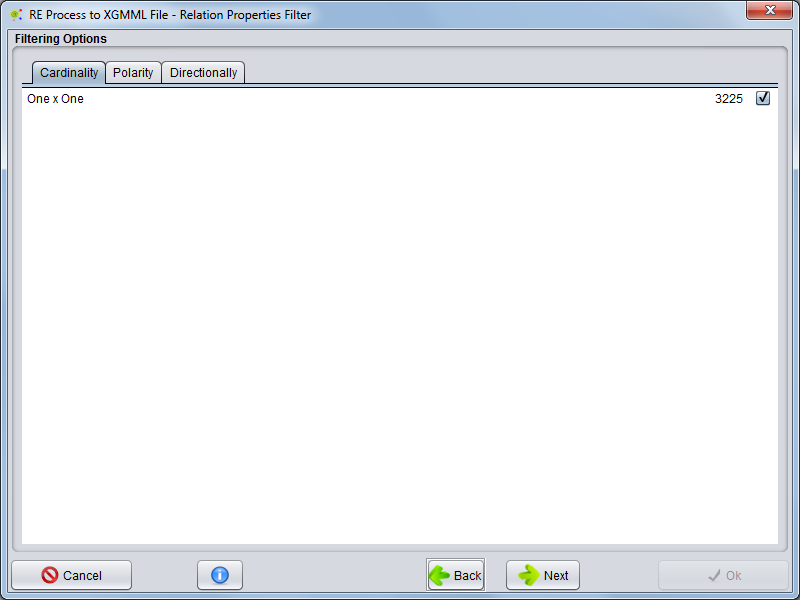
| − | Selecting Cardinality tab | + | Selecting the Cardinality tab, you can select which types of cardinalities to include in the network. |
| + | |||
[[File:RE_To_XGMML_3a.png|center|800px]] | [[File:RE_To_XGMML_3a.png|center|800px]] | ||
| + | |||
=== Polarity === | === Polarity === | ||
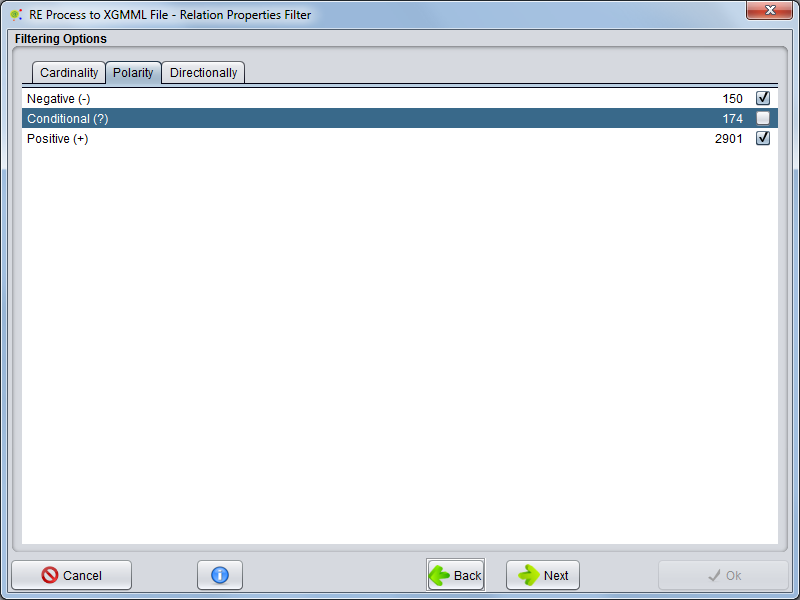
Selecting Polarity tab the user can select witch polarities will be present to form network | Selecting Polarity tab the user can select witch polarities will be present to form network | ||
| + | |||
[[File:RE_To_XGMML_3b.png|center|800px]] | [[File:RE_To_XGMML_3b.png|center|800px]] | ||
| + | |||
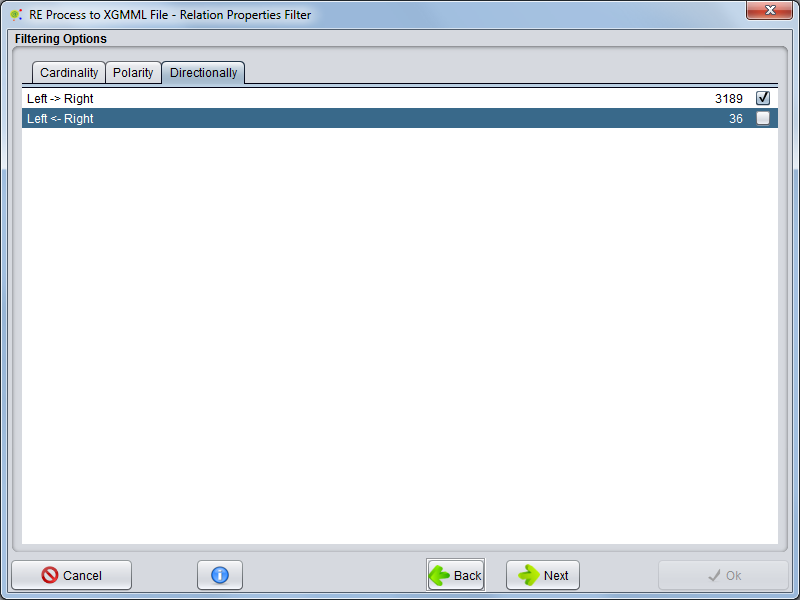
=== Directionally === | === Directionally === | ||
Selecting Directionally tab the user can select witch directionallies will be present to form network | Selecting Directionally tab the user can select witch directionallies will be present to form network | ||
| + | |||
[[File:RE_To_XGMML_3c.png|center|800px]] | [[File:RE_To_XGMML_3c.png|center|800px]] | ||
| + | |||
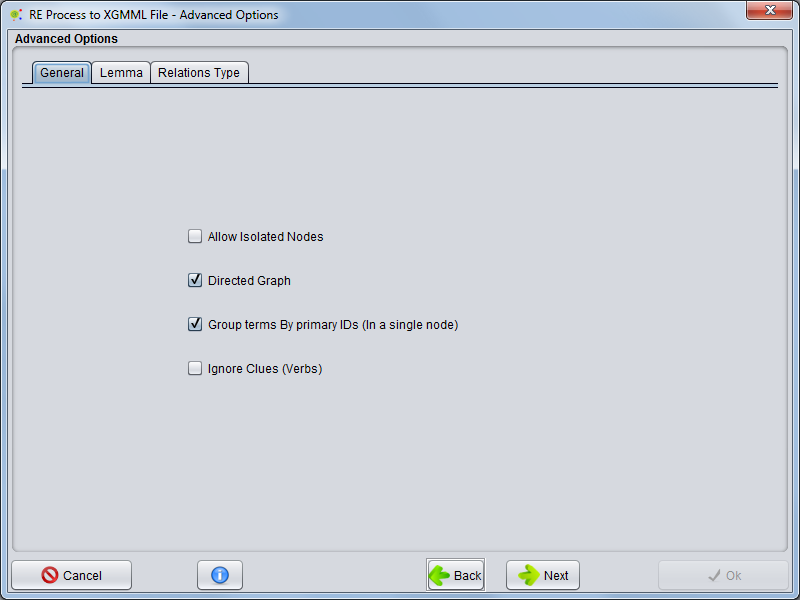
== Advanced Options == | == Advanced Options == | ||
Revision as of 14:37, 27 February 2013
Contents
Operation
This operation allows you to export the entities/relations contained in an REProcess to a network/ graph file in the XGMML format. This file can be afterwards be opened in network visualization and topology analysis such as Cytoscape
To perform this operation, you need to first select the desired RE Process and right click to select Corpus -> RE -> Export To File XGMML
A wizard will be launched for the configuration of the export process.
Entity Selection - Classes (Nodes)
In the first panel, you can choose which entities groups (classes) will be exported to the network file.
Relation Selection
In the next panel, you can filter the relationships to be included in the network to export. Here, you can selecting which features the relationships need to have (e.g. in terms of directionality, cardinality and polarity).
Cardinality
Selecting the Cardinality tab, you can select which types of cardinalities to include in the network.
Polarity
Selecting Polarity tab the user can select witch polarities will be present to form network
Directionally
Selecting Directionally tab the user can select witch directionallies will be present to form network
Advanced Options
Continuing in the next panel the user can choose advanced options
General
Selecting General tab the user can configure some options
- Allow isolated nodes - The user can select whether the network has isolates nodes (Nodes without any connection between other nodes)
- Directed Graph - The user can define whether a graph is directed or not.
- Group Terms By primary ID's (In a single node) - The user can choose whether an entity is a node or a set of interconnected entities (entities and synonyms) are merged into a single node.
- Ignore Clues - The user can choose whether there can be multiple edges between two nodes (using the clues of relationships) or if all clues are ignored and only exist a single connection between two nodes
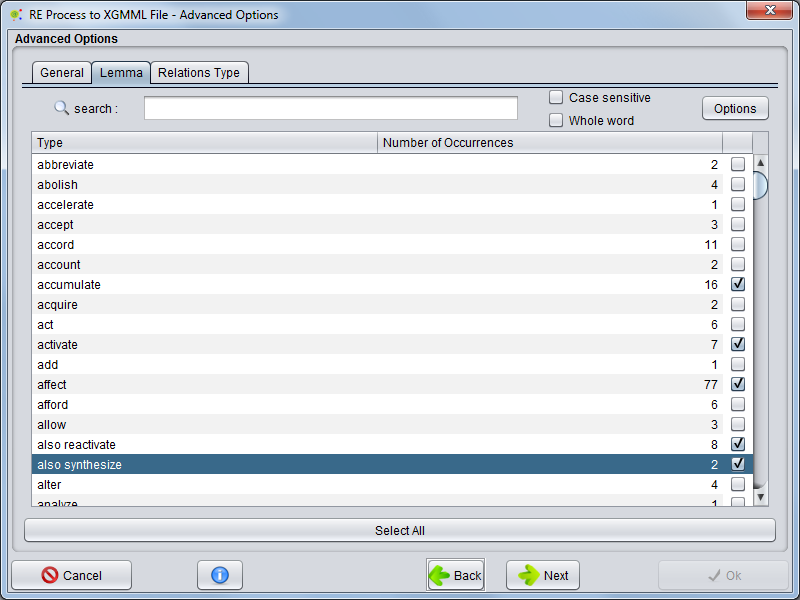
Lemma
Selecting Lemma tab the user can choose witch lemma clues will be present in network edges.
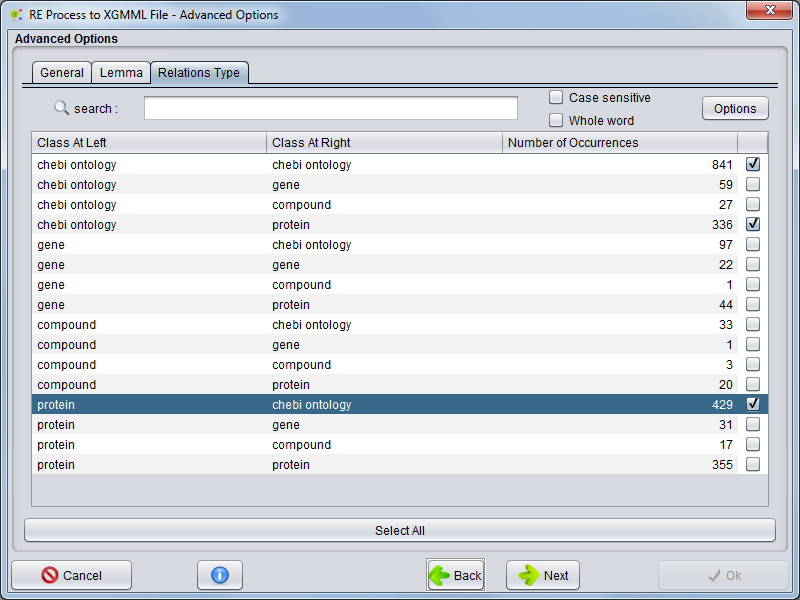
Relation Type
Selecting Relation Type tab the user can choose witch relation types will be present in network edges.
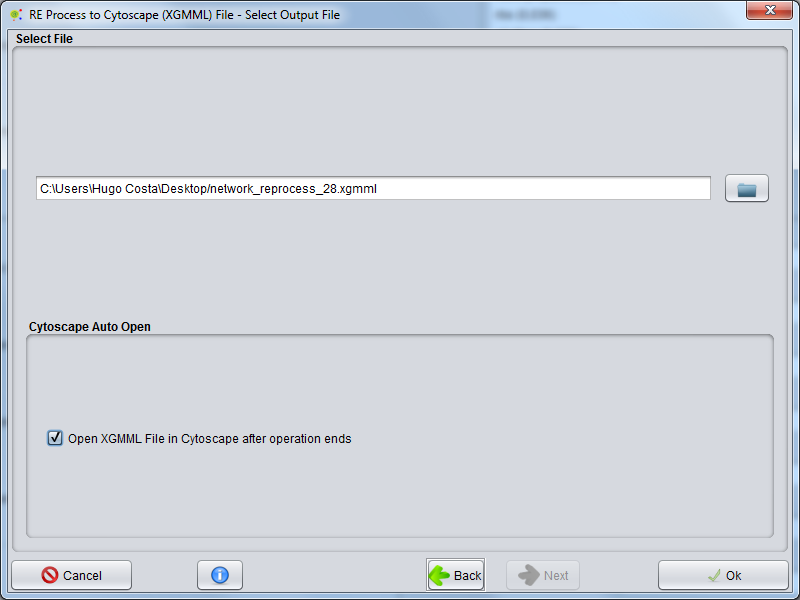
Select File
In final wizard panel the user can choose where to save the network file (.XGMML)
Result
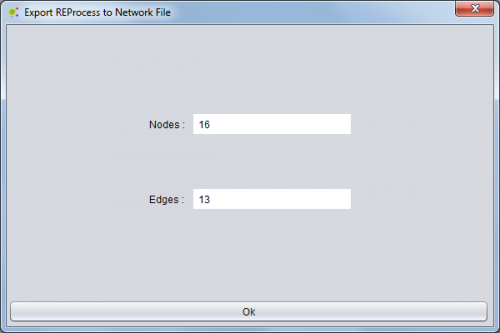
Upon completion of setting a new operation is launched and a time left panel as shown.
After the end of the operation is the user can see an operation report containing information about network exported (number of nodes and edges)
The created file can be opened in tools that have capability to import .XGMML files, such as Cytoscape