Difference between revisions of "NER Model Creation"
Anote2Wiki (talk | contribs) (→New Configuration or Load Configuration) |
Anote2Wiki (talk | contribs) (→Select NLP Tokenizer System) |
||
| Line 18: | Line 18: | ||
[[File:Select_New_or_Load_Configuration_By_BioTML_Tagger.png|center]] | [[File:Select_New_or_Load_Configuration_By_BioTML_Tagger.png|center]] | ||
| + | |||
== Select NLP Tokenizer System == | == Select NLP Tokenizer System == | ||
| − | A GUI is presented to select the possible NLP systems that are integrated in the BioTML framework. Those systems are used to | + | |
| − | The possible NLP systems to be chosen are the ClearNLP, | + | A GUI is presented to select the possible NLP systems that are integrated in the BioTML framework. Those systems are used to perform the tokenization of all documents to create a data matrix for machine learning algorithms.<br /> |
| + | The possible NLP systems to be chosen are the ClearNLP, Stanford Core NLP and OpenNLP. Each system contains a description that is presented on this GUI. | ||
[[File:Select_NLP_System_By_BioTML.png|center]] | [[File:Select_NLP_System_By_BioTML.png|center]] | ||
| − | |||
== BioTML Features Selection == | == BioTML Features Selection == | ||
Revision as of 13:44, 4 August 2015
Contents
Select Option
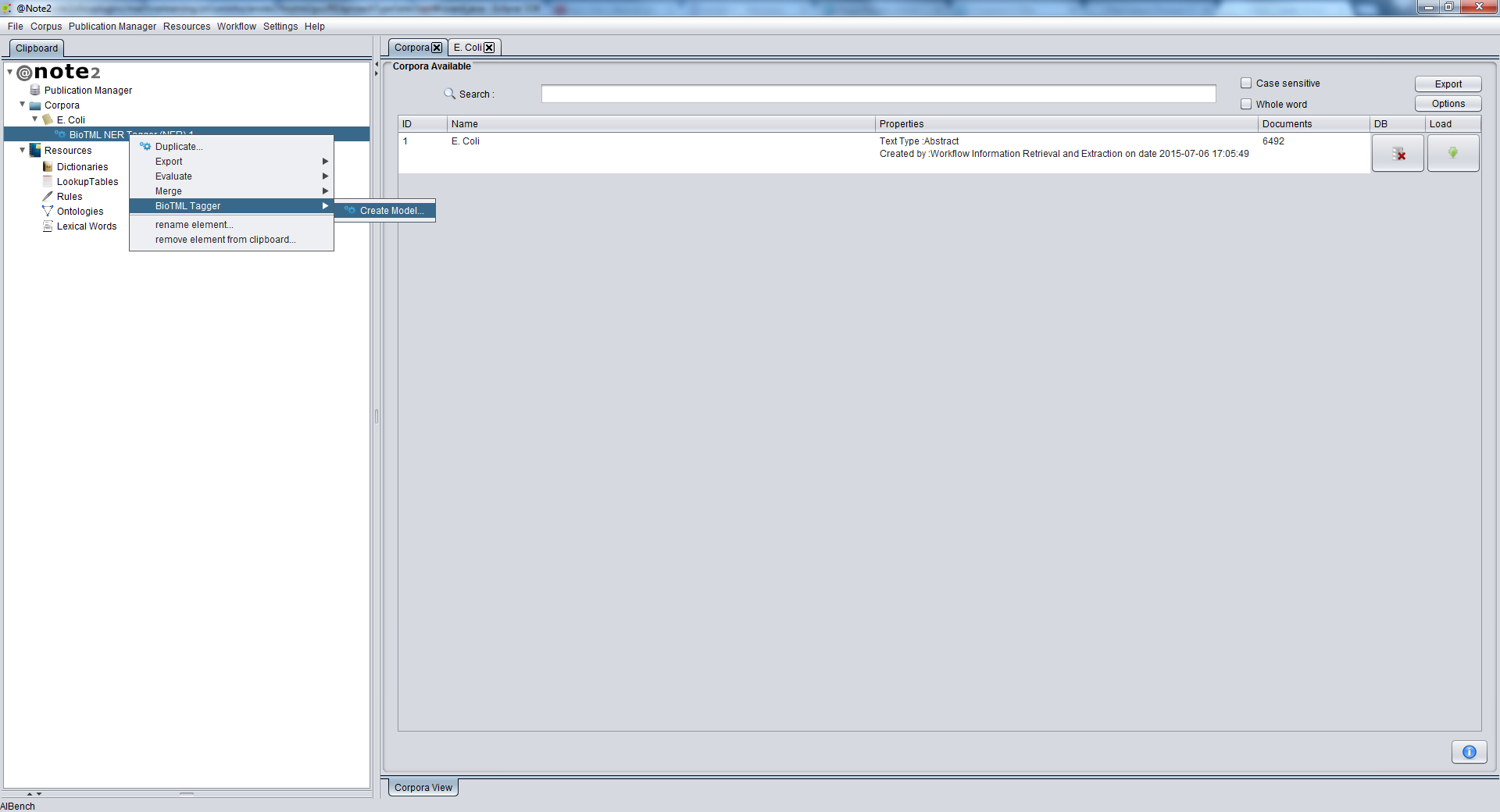
To create an NER Machine Learning Model using BioTML, the first step is to select the corpus with the NER annotations.
To do this, start by loading the desired NER Process to the Clipboard.
Selecting the NER Process, you should right click it and choose NER Process -> BioTML Tagger -> Create Model
New Configuration or Load Configuration
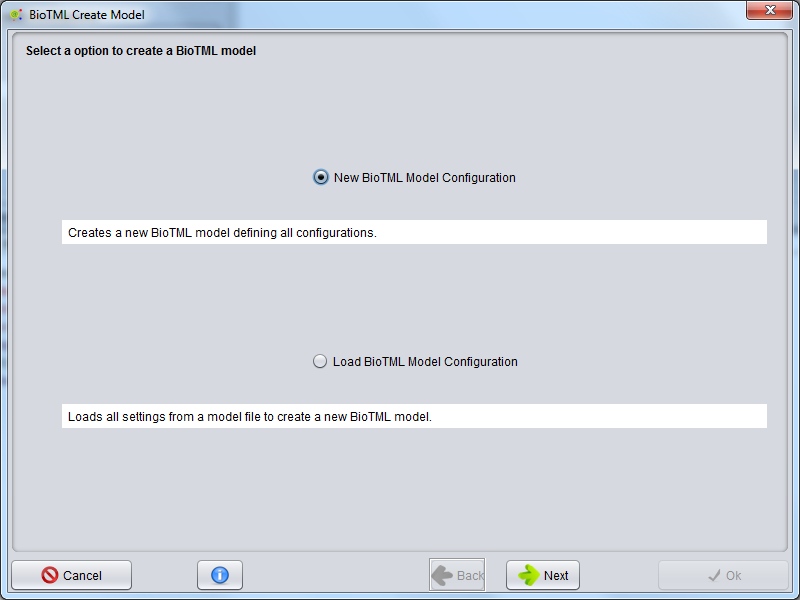
A wizard will be presented to configure the model creation process. The first step allows to select two options: Create New BioTML Model Configuration or Load BioTML Model Configuration.
To start a new model configuration, select Create New BioTML Model Configuration and press the Next button.
Select NLP Tokenizer System
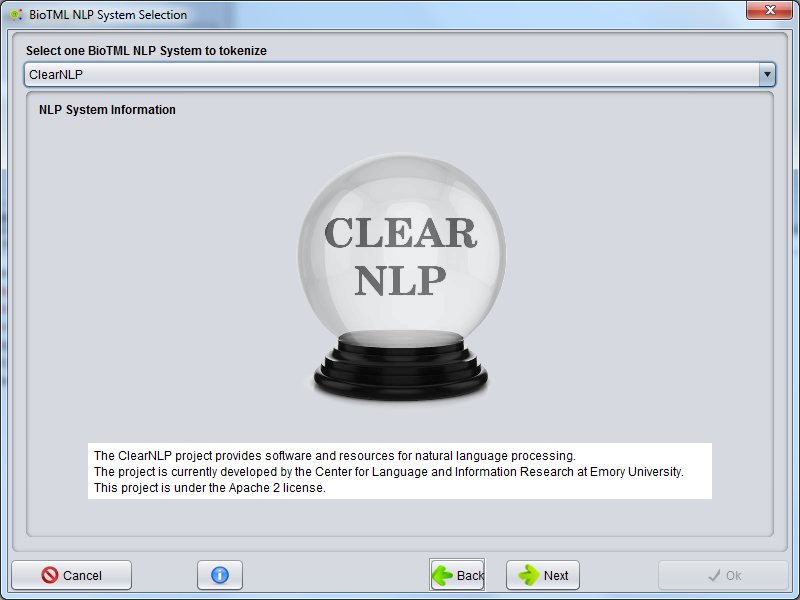
A GUI is presented to select the possible NLP systems that are integrated in the BioTML framework. Those systems are used to perform the tokenization of all documents to create a data matrix for machine learning algorithms.
The possible NLP systems to be chosen are the ClearNLP, Stanford Core NLP and OpenNLP. Each system contains a description that is presented on this GUI.
BioTML Features Selection
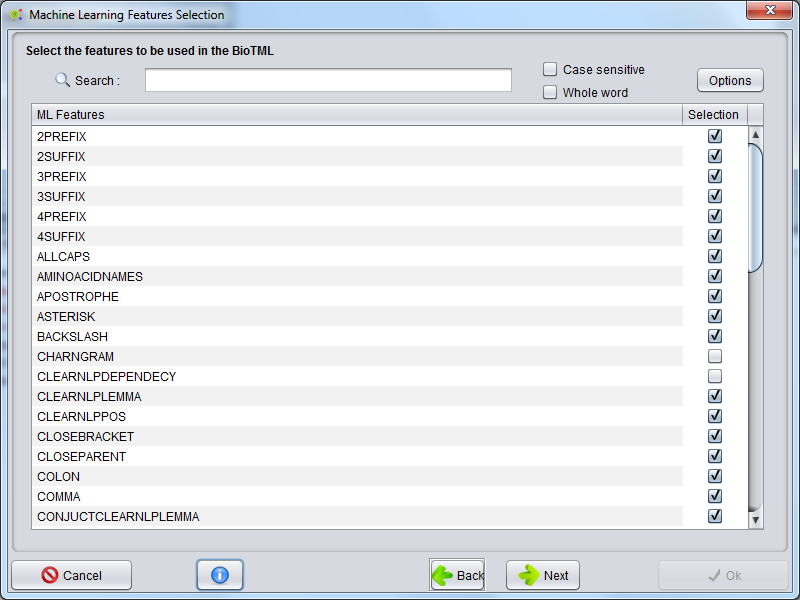
The features selection for machine learning data matrix are selected in this GUI. Regarding the number and type of features, the produced model could have more or less fitting into the data. This selection will have a great impact in the prediction capability, recall and accuracy of the model during the NER annotation.
Attention: The number of features and some feature types could dramatically increase the memory and CPU usage!!
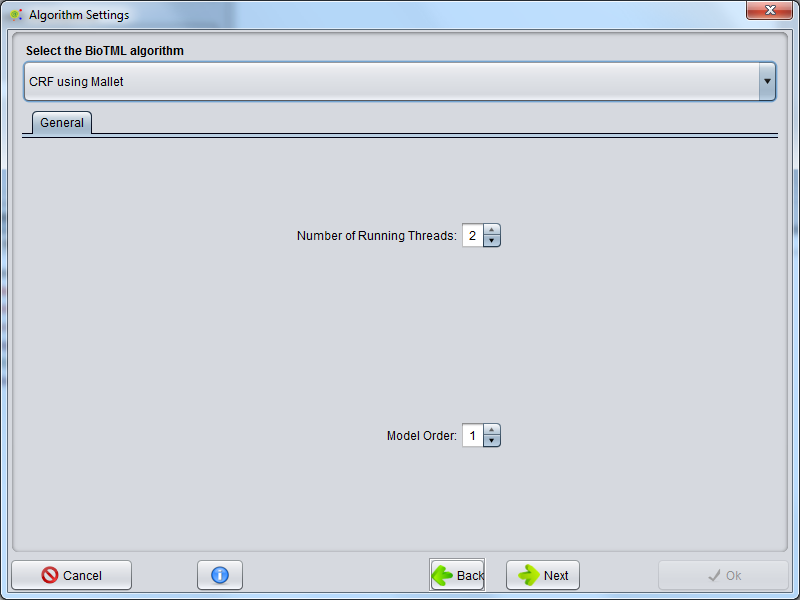
BioTML Model Algorithm Configuration
The machine learning algorithm is selected in this GUI. Advanced settings could appear regarding the selected algorithm type.
For further information's about those advanced configurations please visit the urls: For CRF Informations or For SVM informations
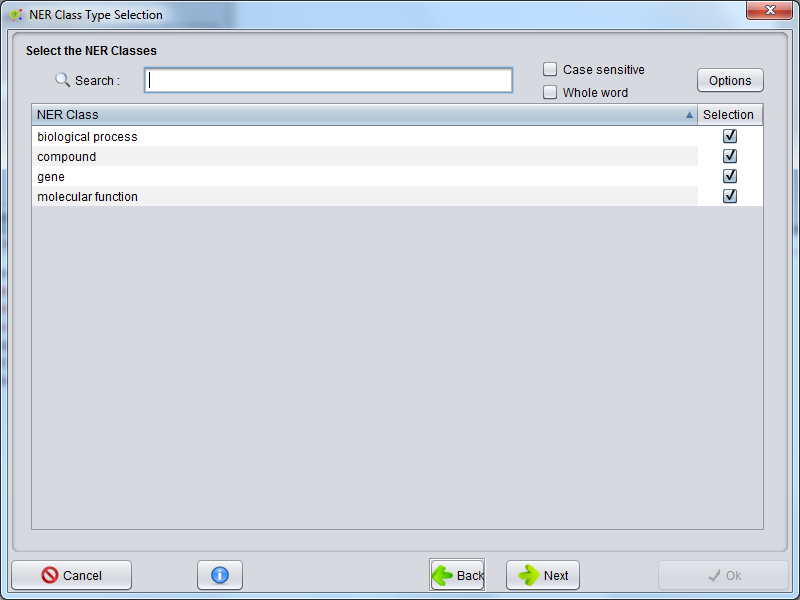
Select_NER_Classes_For_BioTML_Model_Creation
After the algorithm selection, a GUI is presented to select the possible NER classes that will be used to train the model.
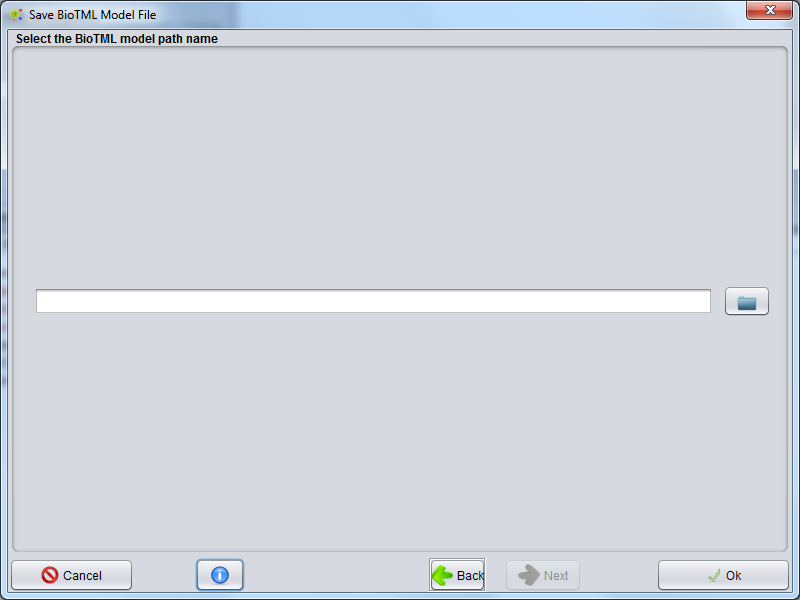
Save_BioTML_Model_File
All configurations for NER Model creation are defined. In this Gui, you could define the directory or Zip file that will store the NER Model.
Result
The NER model creation operation will start and a working window is shown, indicating the execution of the operation. The NER model creation operation will take a few minutes or hours, depending on corpus size and model configurations. When the process ends, a new Zip file containing the NER model will be added to the defined directory in the last GUI.