Corpus Relation Extraction By BioTML Tagger
Contents
Operation
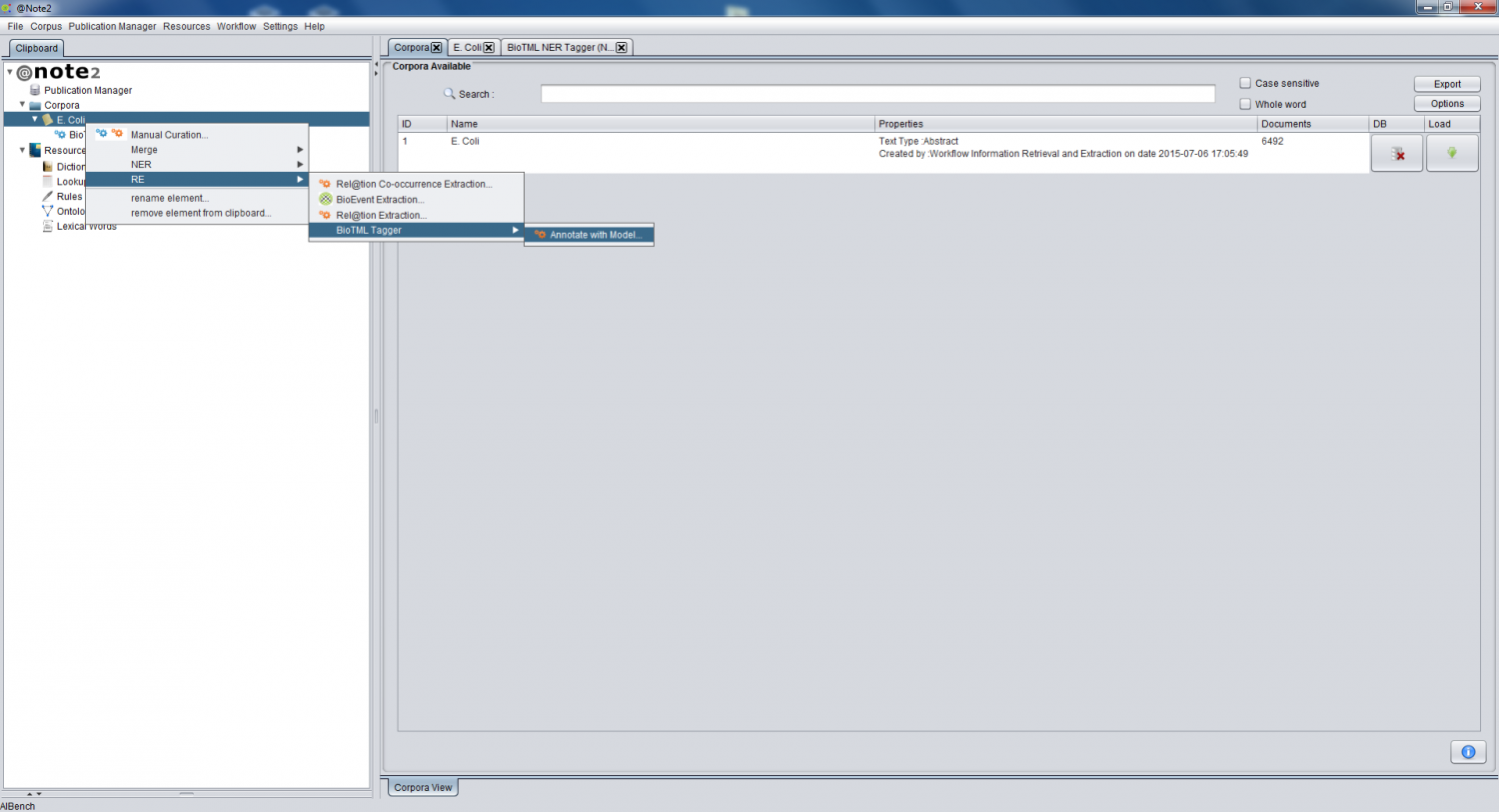
To perform a Relation Extraction By BioTML Tagger (RE) process based in BioTML RE model, you should right click a Corpus data-type object and select the option Corpus -> RE-> BioTML Tagger -> Annotate with Model. If the Corpus is not in the clipboard, you should begin by loading the Corpus to the Clipboard.
A new wizard is presented that allows to configure the RE process.
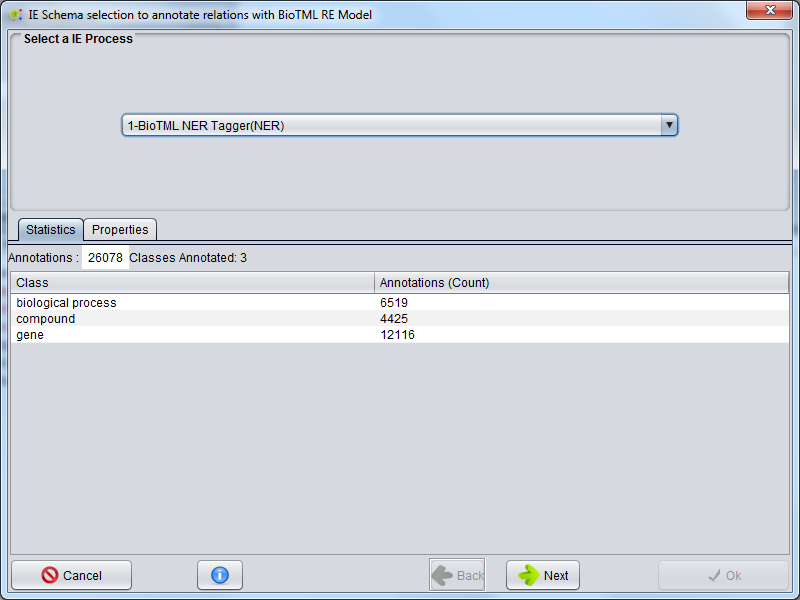
Entity Process Selection For BioTML RE Model Annotation
The first panel enables the selection of the processes that contain the entities annotated, allowing to view some statistics and process properties. After selecting the process press Next to continue.
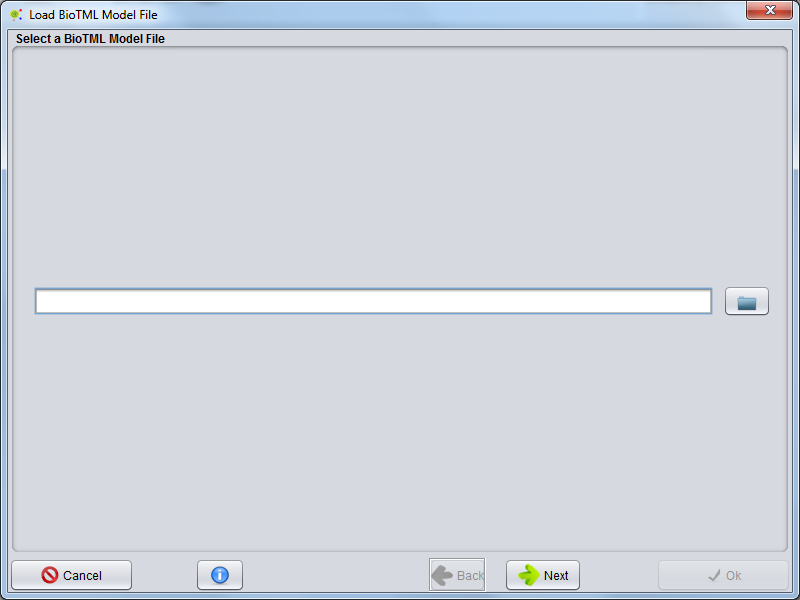
Load_BioTML_Model_File_For_Annotation
A GUI appears allowing to select an BioTML RE model file. Select a compatible BioTML model file (could be a .zip or a .gz file) and press next. The BioTML Model loading will start.
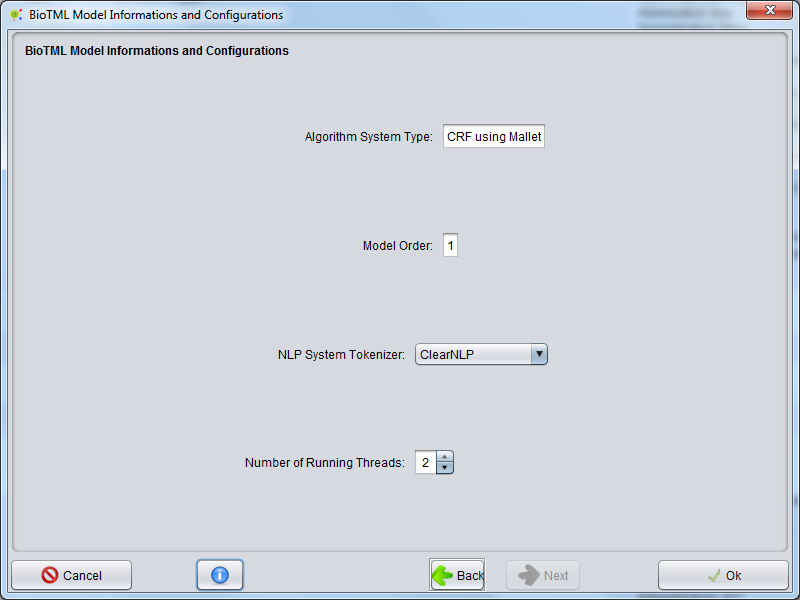
Select BioTML Model Configurations To Annotate
The selected model will contains RE algorithm and configurations.
The RE BioTML Tagger allows you to define options for annotations, as follows:
In the NLP System Tokenizer option, you could select the natural language processing system used for tokenization of all documents in order to annotate.
In the Number of running Threads option, you can select the number of processing threads that the RE BioTML Tagger algorithm will be able to use.
Result
The RE operation will start and a progress window is shown, indicating the execution of the operation. It will take a few minutes or hours, depending on corpus size, number of given threads and features used automatically by BioTML model. When the process ends, a new RE Process object will be added to the Corpus Process View.