Difference between revisions of "Corpus Create Annotation Schema By Chemical Tagger"
Anote2Wiki (talk | contribs) |
|||
| (5 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
One of the options for performing NER is the use of the Chemical Tagger NER algorithm (for more details please visit http://gate.ac.uk/sale/tao/splitch21.html#sec:parsers:chemistrytagger ). | One of the options for performing NER is the use of the Chemical Tagger NER algorithm (for more details please visit http://gate.ac.uk/sale/tao/splitch21.html#sec:parsers:chemistrytagger ). | ||
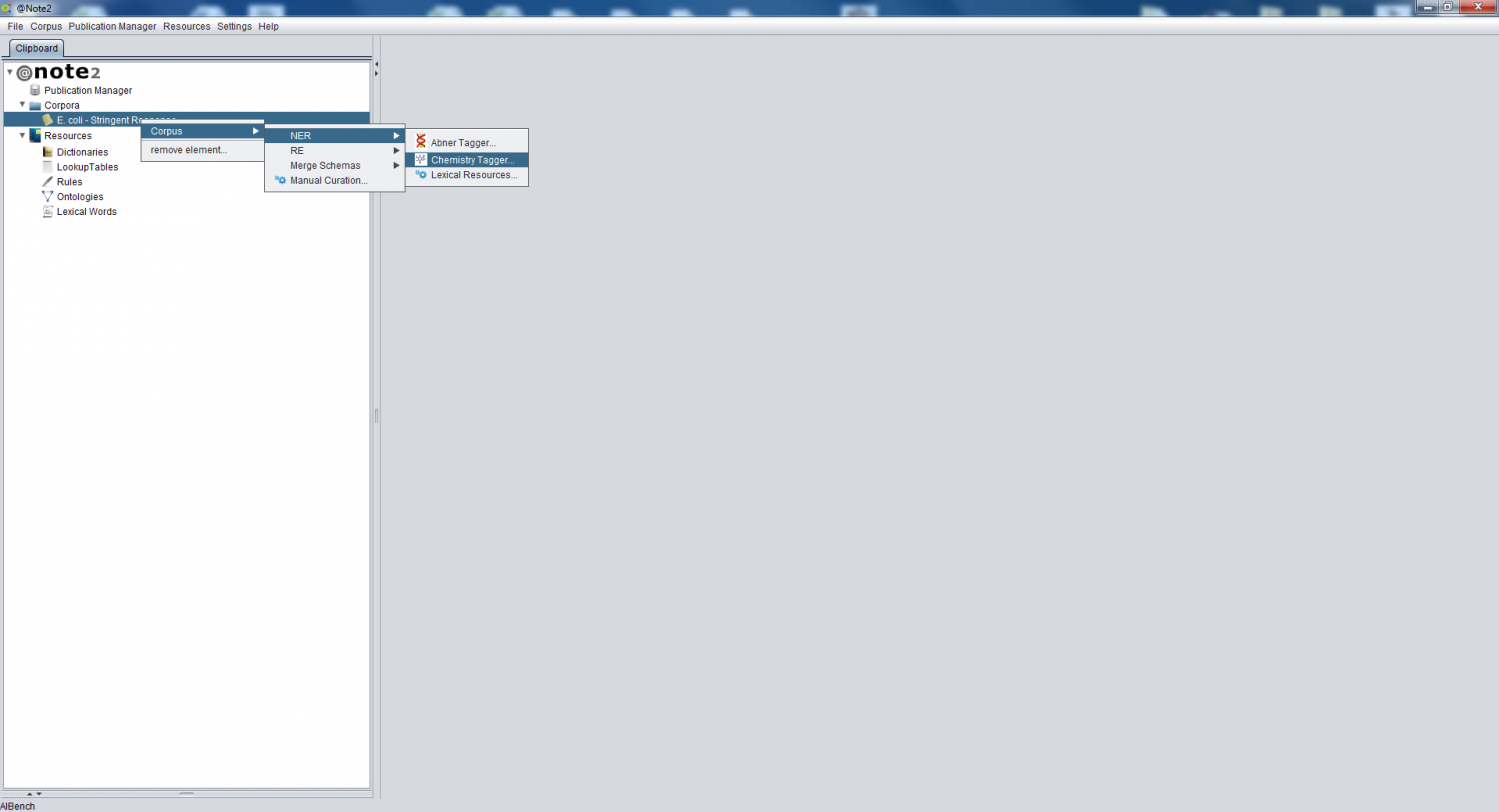
| − | Selecting the Corpus object on the clipboard, you should right click over it an choose '''Corpus -> NER -> | + | Selecting the Corpus object on the clipboard, you should right click over it an choose '''Corpus -> NER -> Chemistry Tagger''' |
| + | |||
[[Image:Corpus_Process_NER_Chemical_Tagger.png|1500px|center]] | [[Image:Corpus_Process_NER_Chemical_Tagger.png|1500px|center]] | ||
| + | |||
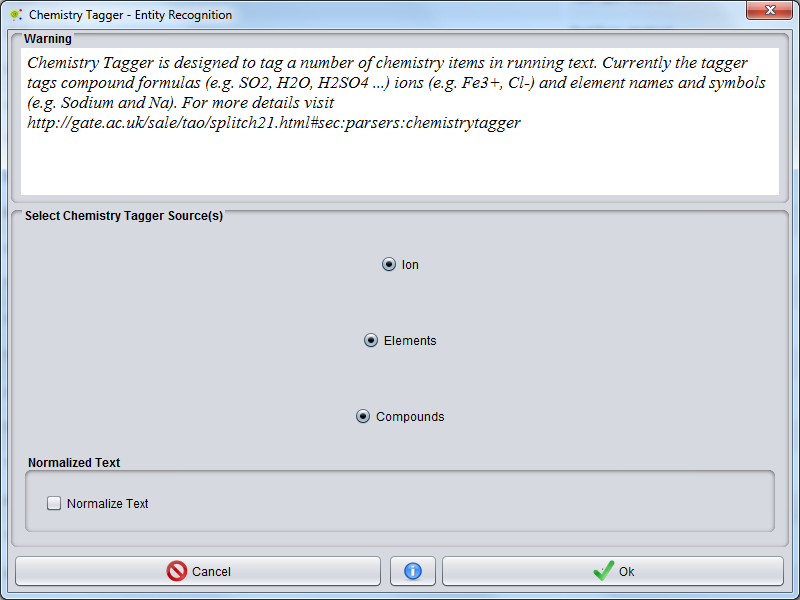
A GUI will be presented, where you can select tagger sources: | A GUI will be presented, where you can select tagger sources: | ||
| − | |||
| − | |||
*'''Ion'''(e.g. Fe3+, Cl-). | *'''Ion'''(e.g. Fe3+, Cl-). | ||
| Line 16: | Line 16: | ||
*'''Element''' names and symbols (e.g. Sodium and Na). | *'''Element''' names and symbols (e.g. Sodium and Na). | ||
| + | |||
| + | |||
| + | [[Image:NER_ChemicalTagger1.png|800px|center]] | ||
| + | |||
| + | '''Normalization option''' allow to add white space between words delimiters and increase the entity recognition accuracy. | ||
| + | |||
Pressing '''Ok''' the operation will be launched and a progress window will be shown, indicating the execution of the operation and estimate time left. | Pressing '''Ok''' the operation will be launched and a progress window will be shown, indicating the execution of the operation and estimate time left. | ||
| − | The NER operation will take a few minutes or hours depending on the number of documents | + | The NER operation will take a few minutes or hours depending on the number of documents and document size. |
| − | When the process | + | When the process ends, a new '''NER Process''' object will be added to the clipboard and can be visualized through the [[Corpora_Load_Corpus|''Corpus Process View'']]. |
Latest revision as of 16:54, 8 May 2013
One of the options for performing NER is the use of the Chemical Tagger NER algorithm (for more details please visit http://gate.ac.uk/sale/tao/splitch21.html#sec:parsers:chemistrytagger ).
Selecting the Corpus object on the clipboard, you should right click over it an choose Corpus -> NER -> Chemistry Tagger
A GUI will be presented, where you can select tagger sources:
- Ion(e.g. Fe3+, Cl-).
- Compound formulas (e.g. SO2, H2O, H2SO4 ...).
- Element names and symbols (e.g. Sodium and Na).
Normalization option allow to add white space between words delimiters and increase the entity recognition accuracy.
Pressing Ok the operation will be launched and a progress window will be shown, indicating the execution of the operation and estimate time left.
The NER operation will take a few minutes or hours depending on the number of documents and document size.
When the process ends, a new NER Process object will be added to the clipboard and can be visualized through the Corpus Process View.