Difference between revisions of "Corpus Create Annotation Schema By Chemical Tagger"
From Anote2Wiki
Anote2Wiki (talk | contribs) |
Anote2Wiki (talk | contribs) |
||
| Line 7: | Line 7: | ||
[[Image:Corpus_Process_NER_Chemical_Tagger.png|1500px|center]] | [[Image:Corpus_Process_NER_Chemical_Tagger.png|1500px|center]] | ||
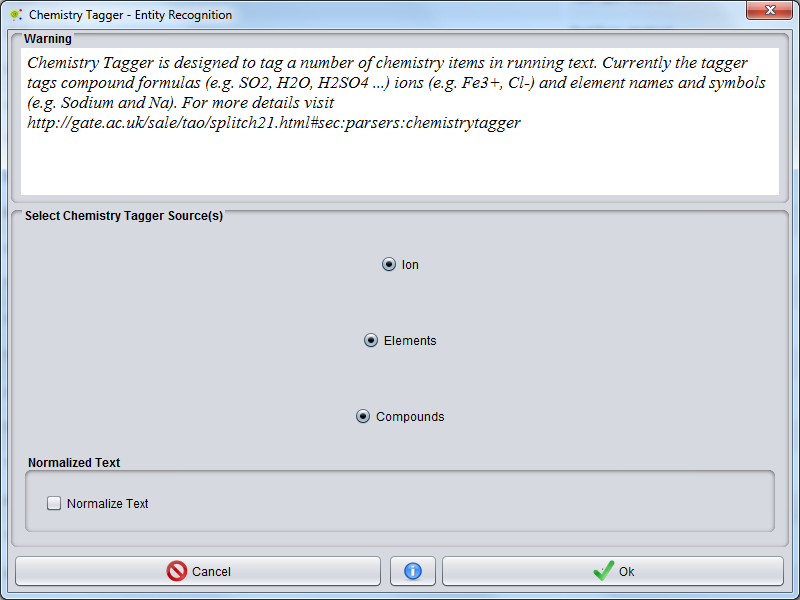
| − | A GUI will be presented, where you can select | + | A GUI will be presented, where you can select tagger sources: |
[[Image:NER_ChemicalTagger1.png|800px|center]] | [[Image:NER_ChemicalTagger1.png|800px|center]] | ||
Revision as of 00:31, 16 January 2013
One of the options for performing NER is the use of the Chemical Tagger NER algorithm (for more details please visit http://gate.ac.uk/sale/tao/splitch21.html#sec:parsers:chemistrytagger ).
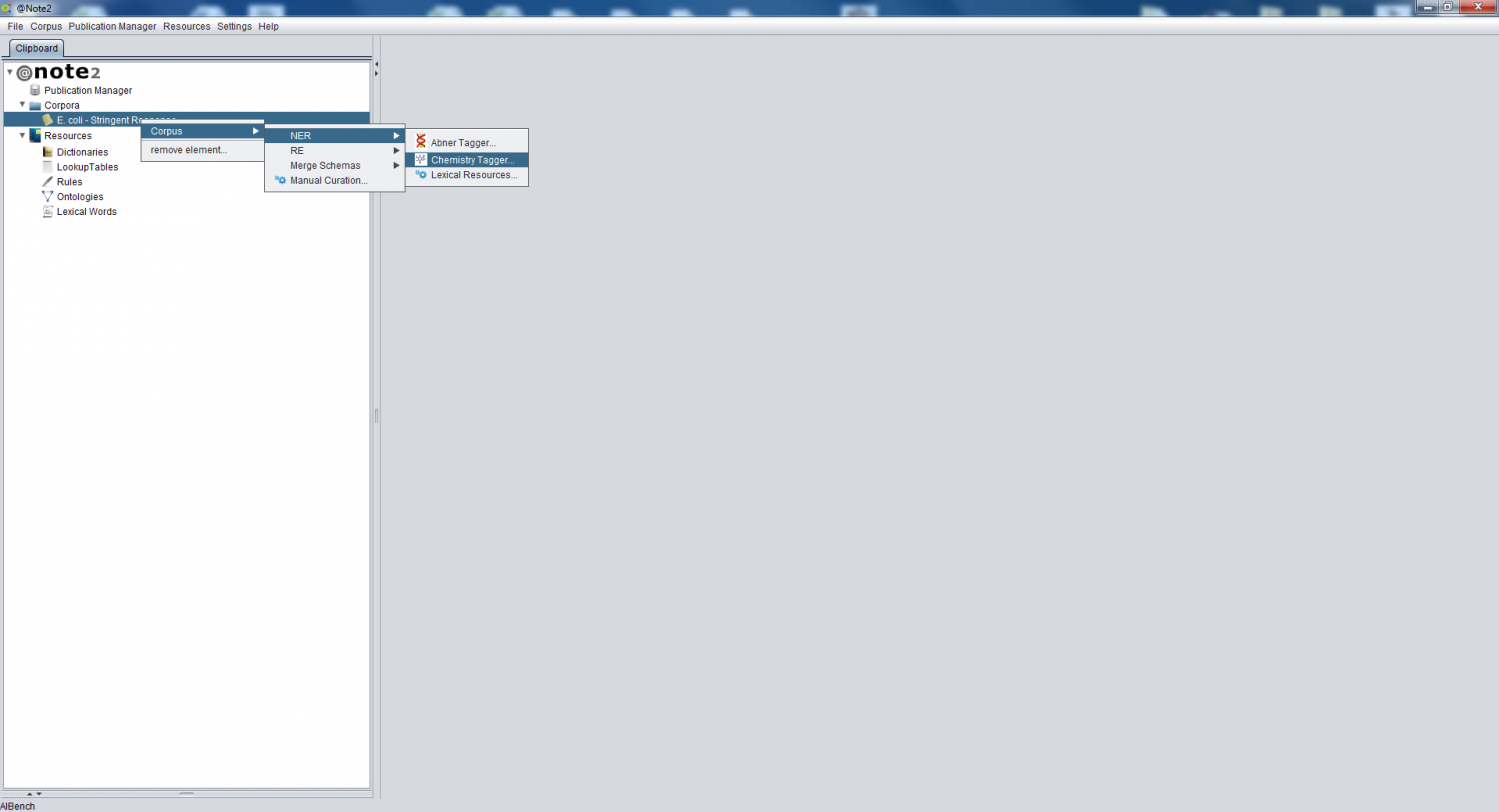
Selecting the Corpus object on the clipboard, you should right click over it an choose Corpus -> NER -> Chemical Tagger
A GUI will be presented, where you can select tagger sources:
- Ion(e.g. Fe3+, Cl-).
- Compound formulas (e.g. SO2, H2O, H2SO4 ...).
- Element names and symbols (e.g. Sodium and Na).
Pressing Ok the operation will be launched and a progress window will be shown, indicating the execution of the operation and estimate time left. The NER operation will take a few minutes or hours depending on the number of documents, document size and used resources.
When the process terminates, a new NER Process object will be added to the Corpus Process View.